0 Genotype By Sequencing
Genotype by Sequencing (GBS): is a molecular technique that allows genomewide genotyping of a population of organisms by performing reduced representation sequencing of their genome. GBS is a robust, simple and affordable means of SNP discovery and mapping. Users of GBS often aim to find Single Nucleotide Polymorphisms (SNPs) within a population to: promote understanding of the relationships between the organisms within, or utilize the genotyping information for genomic selection, prediction or trait analysis.
0.1 Methods
Generating a reduced representation of a genome requires using restriction endonucleases to cleave the double stranded genomic DNA at any position the enzymes unique cut site is found. By utilizing one or more restriction endonucleases, the genomic DNA can be cleaved into manageable fragments. Utilizing different restriction enzymes or combinations of restriction enzyme(s) allow a GBS user to sequence different sites throughout the genome. After enzymatic cleavage the sticky ends of DNA that are left freely floating are used as anchors for the ligation of barcoded adaptors.
The barcoded adaptors serve several important functions:
- A way to indentify each sample uniquely from the others in pooled DNA sequencing
- A DNA template to use for PCR amplification of the DNA before sequencing
- A structural template to adhere other sequencing instrument specific adaptors to
After barcode ligation the DNA is amplified via PCR, purified and quantified before sequencing. Upon sequencing, generally many samples are pooled into a sequencing lane to provide cost effective sharing of the massive sequencing depth provided by todays sequencing instruments.
Your sequencing data has been produced on our NovaSeq 6000. Information about sequencing density, speed and accuracy for this instrument can be found here.
1 Sample Definitions
1.1 Input Data
Specific information pertaining to the input data for your project is shown in Table 1 below.
| Date: | 01/09/2020 |
|---|---|
| Raw Data Path: | /mnt/grl/brc/data/Client/0190620/V2 |
| Genome Path: | /mnt/genomes/Spinacia_oleracea/primary.fa |
| Enzyme: | ApeKI |
The deliverable directory contains numerous files consisting of alignment maps, variant calls, and quality control metrics. These files contain the core information from your project. Content description and explanations of these files are given in the appropriate sections below. Many of these files contain metadata which contain additional information which we do not explicitly pull into this report. Information such as the sequencing flowcell, instrument ID, and cluster are embedded in the names of the sequence alignments.
1.2 Fastq Format
Your raw GBS data can be found in the file in the FASTQ format. The Illumina file naming structure is traditionally:
{Sample_Identifier}_{Sample_Number}_{Lane}_{Read_1_or_Read_2}_001.fastq.gz
A random sample might be named:
Plate-2-Maize_S2_L004_R1_001.fastq.gz
After decompression, FASTQ files have four lines per sequence:
- Line 1: begins with a '@' character and is followed by a sequence identifier and an optional description.
- Line 2: is the raw sequence letters.
- Line 3: begins with a '+' character and is optionally followed by the same sequence identifier (and any description) again.
- Line 4: encodes the quality values for the sequence in Line 2, and must contain the same number of symbols as letters in the sequence.
For Illumina FASTQ files the sequence identifier contains information that can be used to identify metadata about the run.
@<instrument>:<run number>:<flowcell ID>:<lane>:<tile>:<x-pos>:<y-pos> <read>:<is filtered>:<control number>:<sample number>
| Element | Requirements | Description |
|---|---|---|
| @ | @ | Each sequence identifier line starts with @ |
| <instrument> | Characters allowed: a–z, A–Z, 0–9, _ | Instrument ID |
| <run number> | Numerical | Run number on instrument |
| <flowcell ID> | Characters allowed: a–z, A–Z, 0–9 | A Unique string of characters that identifies the flowcell. |
| <lane> | Numerical | Lane number |
| <tile> | Numerical | Tile number |
| <x_pos> | Numerical | X coordinate of cluster |
| <y_pos> | Numerical | Y coordinate of cluster |
| <read> | Numerical | Read number. 1 can be single read or Read 2 of paired-end |
| <is filtered> | Y or N | Y if the read is filtered (did not pass), N otherwise |
| <control number> | Numerical | 0 when none of the control bits are on, otherwise it is an even number. |
| <sample number> | Numerical | Sample number from sample sheet |
2 Quality Control
Raw sequence data may contain residual sequence that is artificially introduced as a result of the laboratory methods used to prepare the DNA for sequencing. These sequence artifacts may introduce bias an increase the complexity of downstream data processing. Additionally, sequencing quality can be affected by various factors such as variation in chemical reagents, instrument platform, temperature or humidity changes, initial DNA quality or concentration. Due to the imperfect nature of the sequencing process and limitations of the optical instruments (batch effects, sequencing error, GC bias, duplicate reads, copy number variation, mapability), base calling has inherent uncertainty associated with it. The magnitude of uncertainty in each base call is represented by an error probability or Phred (pronounced like “fred”) score. This parameter is denoted formally as \(Q\) and is proportional to the probability \(p\) that a base call is incorrect, where \(Q = -10log~10~(p)\).
Any biological inference or conclusion depends on the overall quality of your data. Therfore a quality control (QC) step is essential for providing high quality results. The following QC metrics calculated from your data are designed to help you evaluate the overall technical quality of your experiment. You should always review these metrics to ensure your data are of good quality and lack obvious problems or biases, which may affect how you can ultimately use these data.
2.1 Read Trimming
During sequencing library construction, short oligonucleotides are ligated to the ends DNA fragments to be sequenced, so that they can be combined with primers for PCR amplification. While sequencing the fragments, if the read length is greater than that of the target DNA, the adapter sequence next to the unknown DNA sequence of interest is also sequenced, sometimes only partially. To recover the true target DNA sequence, it is required to identify the adapter sequence and remove it. Moreover, to improve further the quality of the sequence reads, skewer also trims the 3’ end of the fragment until a Phred quality of 20 is reached. This is equivalent to a base call accuracy of 99%. More information regarding Phred scores can be obtained from (Ewing et al. 1998) and in (Ewing & Green 1998).
The Bioinformatics Resource Center performs a quality control step to remove any sequencing adapters and low quality bases. The trimming software skewer (Jiang et al. 2014) was used to pre-process raw fastq files. Skewer implements an efficient dynamic programming algorithm designed to remove adapters and/or primers from sequence reads. Reads that are trimmed too short to be of usable length are discarded from downstream analysis along with the corresponding read pair, in the case of paired-end sequencing.
3 Tassel GBS Pipeline Version 2
The Tassel Version 2 GBS pipeline is an extension of the Java program Tassel. Tassel GBS is a scalable high throughput GBS data analysis platform with moderate computing resource requirements (Glaubitz et al. 2014).
The new pipeline stores data to an embedded SQLite database. All steps of the pipeline either read from or write to this database. It is initially created in the GBSSeqToTagDBPlugin step. When running this pipeline, each subsequent step utilizes/adds data from the database created in the first step. A diagram of the database is presented below.
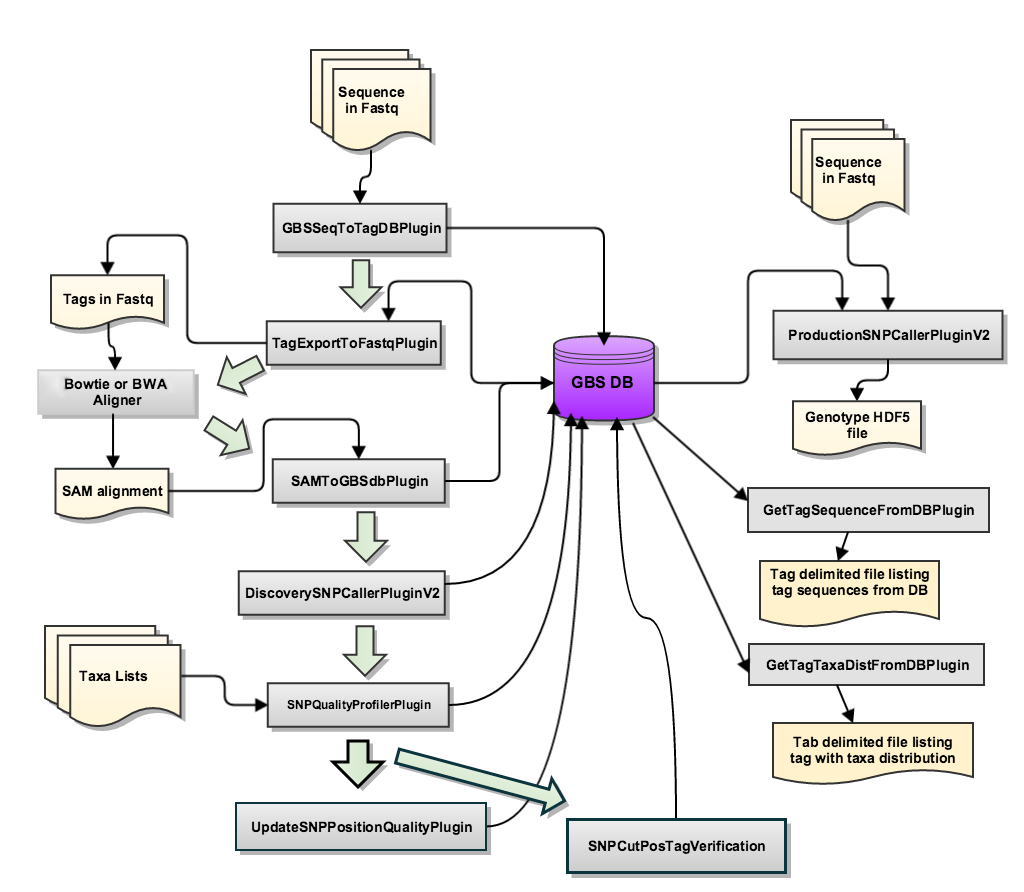
image credit: Tassel GBS v2 Bitbucket
3.1 GBSSeqtoTagDBPlugin
GBSSeqToTagDBPlugin takes fastq files as input, identifies tags and the taxa in which they appear, and stores this data to a local database. It keeps only good reads having a barcode and a cut site and no N’s in the useful part of the sequence. It trims off the barcodes and truncates sequences that (1) have a second cut site, or (2) read into the common adapter.
See the table below for a list of your input files and their associated raw and good barcoded reads:
| Fastq File | Total Raw Reads | Good Barcoded Reads | % Good Barcoded Reads |
|---|---|---|---|
| BHKT72DMXXS24_2_fastq.gz | 328727547 | 314900055 | 95.794 |
| BHKT72DMXXS25_2_fastq.gz | 346133972 | 337261358 | 97.437 |
Raw Reads: is the unprocessed reads within the file before any analysis.
Good Barcoded Reads: are sequence reads with a perfect match to one of the barcodes provided in a barcode key file and with no N’s in the sequence following the barcode up to the trim length.
See the table below to determine the read representation per sample:
| Sample Name | Demultiplexed Reads |
|---|---|
| TB01:201906241500210 | 4871283 |
| TB02:201906241500211 | 3840882 |
| TB03:201906241500212 | 3141617 |
| TB04:201906241500213 | 1220516 |
| TB05:201906241500214 | 403584 |
| TB06:201906241500215 | 3332922 |
| TB07:201906241500216 | 3356085 |
| TB09:201906241500218 | 3206050 |
| TB10:201906241500219 | 1763673 |
| TB11:2019062415002110 | 2537969 |
| TB12:2019062415002111 | 3643434 |
| TB13:2019062415002112 | 904588 |
| TB14:2019062415002113 | 1922513 |
| TB15:2019062415002114 | 3311405 |
| TB16:2019062415002115 | 2159178 |
| TB17:2019062415002116 | 3736498 |
| TB18:2019062415002117 | 3036391 |
| TB19:2019062415002118 | 1038867 |
| TB20:2019062415002119 | 4383077 |
| TB21:2019062415002120 | 3541881 |
| TB22:2019062415002121 | 2879401 |
| TB23:2019062415002122 | 1966082 |
| TB24:2019062415002123 | 2645742 |
| TB25:2019062415002124 | 1720659 |
| TB269:2019062415002163 | 3996310 |
| TB26:2019062415002125 | 2093917 |
| TB27:2019062415002126 | 4019942 |
| TB281:2019062415002161 | 4297906 |
| TB287:2019062415002142 | 4301044 |
| TB288:2019062415002143 | 1394822 |
| TB28:2019062415002127 | 31361 |
| TB292:2019062415002144 | 4693220 |
| TB293:2019062415002145 | 7144642 |
| TB29:2019062415002128 | 2095624 |
| TB30:2019062415002129 | 1473205 |
| TB311:2019062415002164 | 3242522 |
| TB312:2019062415002165 | 3942637 |
| TB313:2019062415002166 | 4693147 |
| TB314:2019062415002167 | 2697100 |
| TB315:2019062415002168 | 4542589 |
| TB317:2019062415002169 | 1417591 |
| TB318:2019062415002170 | 1249588 |
| TB319:2019062415002171 | 3262468 |
| TB31:2019062415002130 | 1319527 |
| TB320:2019062415002191 | 1927806 |
| TB321:2019062415002172 | 3336375 |
| TB322:2019062415002173 | 4053412 |
| TB323:2019062415002190 | 3570 |
| TB324:2019062415002174 | 2941917 |
| TB32:2019062415002131 | 1803268 |
| TB33:2019062415002132 | 3090496 |
| TB342:2019062415002146 | 3898621 |
| TB344:2019062415002147 | 3892773 |
| TB34:2019062415002133 | 2527338 |
| TB351:2019062415002148 | 3546380 |
| TB352:2019062415002149 | 6602342 |
| TB355:2019062415002192 | 3543571 |
| TB356:2019062415002189 | 1076192 |
| TB359:2019062415002175 | 3780938 |
| TB35:2019062415002134 | 2861343 |
| TB360:2019062415002176 | 3531495 |
| TB361:2019062415002177 | 4536715 |
| TB362:2019062415002178 | 3845707 |
| TB363:2019062415002179 | 4377290 |
| TB364:2019062415002180 | 2443930 |
| TB365:2019062415002181 | 3295198 |
| TB366:2019062415002182 | 5336955 |
| TB367:2019062415002183 | 1017752 |
| TB368:2019062415002184 | 2119291 |
| TB369:2019062415002185 | 3487084 |
| TB36:2019062415002135 | 3289158 |
| TB371:2019062415002186 | 2763307 |
| TB375:2019062415002194 | 3362717 |
| TB37:2019062415002136 | 5446743 |
| TB382:2019062415002193 | 2024661 |
| TB385:2019062415002187 | 2118758 |
| TB386:2019062415002188 | 2522771 |
| TB387:2019062415002150 | 3041348 |
| TB388:2019062415002151 | 4596016 |
| TB389:2019062415002152 | 3734361 |
| TB38:2019062415002137 | 3180931 |
| TB390:2019062415002153 | 5009953 |
| TB391:2019062415002154 | 2882213 |
| TB392:2019062415002155 | 3993264 |
| TB39:2019062415002138 | 3939640 |
| TB400:201906241500210 | 3297436 |
| TB401:201906241500211 | 2450620 |
| TB402:201906241500212 | 2269217 |
| TB403:201906241500213 | 2043302 |
| TB404:201906241500214 | 1934339 |
| TB405:201906241500215 | 1620578 |
| TB406:201906241500216 | 3072910 |
| TB407:201906241500217 | 3949526 |
| TB408:201906241500218 | 2728633 |
| TB409:201906241500219 | 3019801 |
| TB40:2019062415002139 | 5146874 |
| TB410:2019062415002110 | 3293726 |
| TB411:2019062415002111 | 1875492 |
| TB412:2019062415002112 | 3249439 |
| TB413:2019062415002113 | 2913555 |
| TB414:2019062415002114 | 2071057 |
| TB415:2019062415002115 | 1421149 |
| TB416:2019062415002116 | 2978207 |
| TB417:2019062415002117 | 2778422 |
| TB418:2019062415002118 | 3286214 |
| TB419:2019062415002119 | 3634284 |
| TB41:2019062415002140 | 3604561 |
| TB420:2019062415002120 | 3569161 |
| TB421:2019062415002121 | 2395300 |
| TB422:2019062415002122 | 3024827 |
| TB423:2019062415002123 | 2770340 |
| TB424:2019062415002156 | 6907389 |
| TB425:2019062415002157 | 3920483 |
| TB427:2019062415002158 | 5905164 |
| TB428:2019062415002124 | 2675804 |
| TB429:2019062415002125 | 4837039 |
| TB42:2019062415002141 | 3598196 |
| TB430:2019062415002126 | 4358571 |
| TB431:2019062415002127 | 2892719 |
| TB432:2019062415002128 | 2722583 |
| TB433:2019062415002129 | 2702163 |
| TB434:2019062415002130 | 2393586 |
| TB435:2019062415002131 | 2061431 |
| TB437:2019062415002132 | 2331902 |
| TB438:2019062415002133 | 3008786 |
| TB439:2019062415002134 | 558757 |
| TB43:2019062415002142 | 4395619 |
| TB440:2019062415002135 | 3902577 |
| TB443:2019062415002136 | 6149376 |
| TB444:2019062415002137 | 4628001 |
| TB445:2019062415002138 | 2960481 |
| TB446:2019062415002139 | 5211966 |
| TB447:2019062415002140 | 2340850 |
| TB448:2019062415002141 | 4919473 |
| TB44:2019062415002143 | 3661816 |
| TB451:2019062415002195 | 1051117 |
| TB453:2019062415002159 | 4219761 |
| TB454:2019062415002160 | 4092276 |
| TB45:2019062415002144 | 4168867 |
| TB46:2019062415002145 | 3281510 |
| TB47:2019062415002146 | 3998183 |
| TB48:2019062415002147 | 3264880 |
| TB49:2019062415002148 | 3983888 |
| TB50:2019062415002149 | 5438900 |
| TB51:2019062415002150 | 4072843 |
| TB52:2019062415002151 | 3902759 |
| TB53:2019062415002152 | 4558727 |
| TB54:2019062415002153 | 4345295 |
| TB55:2019062415002154 | 4012202 |
| TB56:2019062415002155 | 2905396 |
| TB57:2019062415002156 | 3934828 |
| TB58:2019062415002157 | 1416179 |
| TB59:2019062415002158 | 6208739 |
| TB60:2019062415002159 | 2530028 |
| TB61:2019062415002160 | 4539002 |
| TB62:2019062415002161 | 4370473 |
| TB63:2019062415002162 | 4066966 |
| TB64:2019062415002163 | 4761181 |
| TB65:2019062415002164 | 3230965 |
| TB66:2019062415002165 | 3967260 |
| TB67:2019062415002166 | 2896394 |
| TB68:2019062415002167 | 3316771 |
| TB69:2019062415002168 | 1153914 |
| TB70:2019062415002169 | 2232678 |
| TB71:2019062415002170 | 2040634 |
| TB72:2019062415002171 | 2796892 |
| TB73:2019062415002172 | 2094934 |
| TB74:2019062415002173 | 3095355 |
| TB75:2019062415002174 | 3640028 |
| TB76:2019062415002175 | 887970 |
| TB77:2019062415002176 | 1954851 |
| TB78:2019062415002177 | 4320311 |
| TB79:2019062415002178 | 3946669 |
| TB80:2019062415002179 | 3703259 |
| TB81:2019062415002180 | 2535635 |
| TB82:2019062415002181 | 2804720 |
| TB83:2019062415002182 | 3994439 |
| TB84:2019062415002183 | 1980138 |
| TB85:2019062415002184 | 2678242 |
| TB86:2019062415002185 | 3340099 |
| TB87:2019062415002186 | 3009971 |
| TB88:2019062415002187 | 3732187 |
| TB89:2019062415002188 | 3138272 |
| TB90:2019062415002189 | 4309035 |
| TB91:2019062415002190 | 2754301 |
| TB92:2019062415002191 | 2325574 |
| TB93:2019062415002192 | 4015065 |
| TB94:2019062415002193 | 2046390 |
| TB95:2019062415002194 | 3085905 |
| TB96:2019062415002195 | 1710394 |
| blank2:2019062415002162 | 254 |
| blank:201906241500217 | 90212 |
| Total: | 606654479 |
Demultiplexed Reads are sequence reads with a perfect match to one of the barcodes provided in a barcode key file and with no N’s in the sequence following the barcode up to the trim length.
Interpretation: The list above provides the Demultiplexed Reads assigned to each sample. Therefore, the table values should provide a clue as to which samples are most well represented in your GBS experiment (>1x106 Demultiplexed Reads). Low counts (i.e. <1x105) likely indicates sample dropout. Sample dropout occasionally occurs in GBS due to the large number of samples that are pooled on our patterned Illumina flow cells and the error inherent in measuring DNA libraries.
3.2 TagExportToFastqPlugin
TagExportToFastqPlugin retrieves distinct tags stored in the database and reformats them to a FASTQ file that can be read by the Bowtie 2 or BWA aligner program. This plugin leads to one of the main practical advantages of the Tassel GBS pipeline. Namely, the collapse of redundant reads into tags of a predefined tag size. This read collapse drastically shortens the time required for tag/read alignment because of the reduced number of tags compared to the input reads. We utilized 64 bp tags during the analysis of your data. Even a single bp difference in reads of similar sequence will lead to different tags being stored in the database. The output file of TagExportToFastqPlugin is the input to the DNA sequence aligner Bowtie 2.
3.3 GBS Tag Alignment
The BRC uses the Bowtie 2 alignment software for alignment of the FASTQ data
transfered from the TagExportToFastqPlugin. Bowtie 2 is a fast and memory efficient read aligner that supports gapped, local and paired-end alignment modes (Langmead et al. 2012) .
To narrow down the locations a read can map in a large genome, Bowtie 2 extracts multiple "seeds" (smaller DNA sequences) from each
read/tag and aligns them in an ungapped manner to the FM index in an effort to align the entire read. This multiseed heuristic
increases the speed of alignment but can reduce accuracy.
Bowtie 2 deals with inaccuracy of read mapping via two methods:
- Using an alignment scoring scheme which penalizes inaccurate alignments (gaps,insertions and mismatches relative to the reference) and rewards accuracy (correctly matched bases). Simply put, the higher the score the more similar the read/tag is to the reference sequence. If a read's alignment score exceeds the minimum score threshold (adjustable), the read's alignment is considered "valid". The scoring scheme is implemented differently for local vs end-to-end (default) alignments. In the Tassel v2 GBS pipeline we employ the --very-sensitive-local method during mapping of tags to the genome to ensure high accuracy of each tags mapping location.
- Outputting a Mapping Quality (MAPQ) for each read alignment. The MAPQ in Bowtie 2 represents the aligners confidence that the location the read is aligned to is the read's true point of origin. MAPQ ranges from (lowest: 0 --> highest: 42) and is reported as: Q = -10 log10 p, where p is an estimate of the probability that the alignment does not correspond to the read’s true point of origin.
The output of each Bowtie 2 run is a Sequence Alignment Map (SAM) file, the details of which can be found below:
| Col | Field | Description |
|---|---|---|
| 1 | QNAME | Query (pair) NAME |
| 2 | FLAG | bitwise FLAG |
| 3 | RNAME | Reference sequence NAME |
| 4 | POS | 1-based leftmost POSition/coordinate of clipped sequence |
| 5 | MAPQ | MAPping Quality (Phred-scaled) |
| 6 | CIGAR | extended CIGAR string |
| 7 | MRNM | Mate Reference sequence NaMe (‘=’ if same as RNAME) |
| 8 | MPOS | 1-based Mate POSition |
| 9 | ISIZE | Inferred insert SIZE |
| 10 | SEQ | query SEQuence on the same strand as the reference |
| 11 | QUAL | query QUALity (ASCII-33 gives the Phred base quality) |
| 12 | OPT | variable OPTional fields in the format TAG:VTYPE:VALUE |
This data has been provided to you in the GBS_
See below for a table indicating genome mapping of the tags in your population:
| Total Tags | Secondary Tag Alignment | % Secondary Tag Alignment | Supplementary Tag Alignment | % Supplementary Tag Alignment | Mapped Tags | % Mapped Tags |
|---|---|---|---|---|---|---|
| 1175237 | 0 | 0.0 | 0 | 0.0 | 599318 | 50.996 |
3.4 SamToGBSdbPlugin
SAMToGBSdbPlugin reads a SAM file to determine the potential positions of Tags against the reference genome. The plugin updates the current database with information on tag cut positions.
3.5 DiscoverySNPCallerPluginV2
DiscoverySNPCallerPluginV2 takes a GBSv2 database file as input and identifies SNPs from the aligned tags. Tags positioned at the same physical location are aligned against one another, SNPs are called from the aligned tags, and the SNP position and allele data are written to the database.
3.6 SNPQualityProfilerPlugin
This plugin scores all discovered SNPs for various coverage, depth and genotypic statistics for a given set of taxa.
3.7 ProductionSNPCallerPluginV2
This plugin converts data from fastq and keyfile to genotypes, then adds these to a genotype file in VCF or HDF5 format. VCF is the default output.
4 Variant Detection
4.1 Variant Call Format (VCF)
A variant call format (VCF) file is the most common way to communicate a list of variations from the reference genome. VCF is a flexible text based format that is often stored in a gzip compressed format (*.vcf.gz extension). A detailed description of the nuances of this file format can be found at here. Briefly, the format contains meta-information lines, a header line, and then data lines. Each data line is tab-delimited with 8 fixed fields per record, described in the table below. The INFO field is the fixed field that contains a large amount of metadata about the variant. The data contained in the INFO field is variable and will be specified in the VCF header section.
Following the 8 fixed fields, there are optional fields describing the genotype information for each sample (the same types of genotype data must be present for all samples). A FORMAT field is given specifying the order of the colon-separated genotype data for each sample field. See the VCF documentation for more information about genotype fields.
| Col | Field | Description |
|---|---|---|
| 1 | CHROM | Identifies the chromosome on the reference |
| 2 | POS | The reference position, with the 1st base having position 1. |
| 3 | ID | Semi-colon separated list of unique identifiers where available |
| 4 | REF | Reference base(s) |
| 5 | ALT | Comma separated list of alternate non-reference alleles |
| 6 | QUAL | Phred-scaled quality score for the assertion made in ALT |
| 7 | FILTER | A semicolon-separated list of codes for failed filters (PASS or . indicate that the variant passed quality checks) |
| 8 | INFO | Additional information encoded as a semicolon-separated series of short keys with optional values |
| 9 | FORMAT | A colon-separated field that specifies the data types and order for the following <SAMPLE> fields |
| 10+ | <SAMPLE> | The genotype information for the sample specifed in the header line |
The table below provides the number of SNPs that were found per contig:
| Chromosome/Contig | Number of SNPs |
|---|---|
| 1 | 22115 |
| 2 | 21824 |
| 3 | 40542 |
| 4 | 39876 |
| 5 | 21850 |
| 6 | 19540 |
| Total SNPs: | 165747 |
Interpretation: The table above indicates the number of SNPs discovered per Chromosome/Contig. These results indicate the unfiltered SNP marker sites detected by the Tassel v2 GBS pipeline . To generate high quality filtered SNPs you will need to perform traditional site and sample filtering. Depending on your scientific question and organism of interest you may consider following the GATK best practices for variant filtering. A few commonly used variant filtering programs you might consider using are the GUI based program Tassel or the command line programs bcftools, GATK, and vcftools.
5 Sample Relationships
5.1 Principal Component Analysis
Above we used the PCA method to visualize the genetic distance and relatedness of the organisms in the population you provided for GBS. To perform an accurate PCA analysis it is necessary to filter SNP sites, cull weak samples and and impute any missing data. We pre-filter you SNP data for: SNP sites with genotype information at >=60% of the samples in your population. Next, we cull any samples containing inadequate genotype information (<=80% of the pre-filtered SNP sites). Finally, we select only SNP sites containing sufficient genotype data in the population for imputation (i.e. sites with data for >=80% of samples). We then perform whole genome phasing and impute any missing data using the extensively tested Beagle algorithm on these high confidence SNP marker sites before generation of the PCA. During de novo SNP discovery, SNP phasing and imputation are impossible due to the lack of contiguous stretches of variants. Therefore, in de novo analyses, we accept only SNP sites which have data in the entire culled sample population. Information on the number of SNPs and filtering steps used to generate this plot can be found in the ./hapMap/PCA_analysis/Filter.log. Above we have utilized only the first three principal components (which often explain much of a GBS populations genetic variance) for visualization. If you would like further principal component data on your samples, principal components 1-5 calculated by the sklearn PCA function are present in the ./hapMap/PCA_analysis/PCA1.txt file.
Interpreting your PCA:Each point on the PCA represents an individual in your GBS population. If you hover over a point with the mouse, the chart will display the corresponding sample name and principal components (PC1,PC2,PC3 are x, y, and z respectively). Additionally, you can drag the 3D plot to better visualize relationships of the different clusters. Pictures from any angle can be saved by clicking the appropriate options in the upper right corner. Each point is colored based on the unsupervised clustering of the first three principal components. We have optimized the agglomerative clustering of the PCA components in your data by testing the clustering with up to 10 data clusters and picking the optimal number clusters such that: if another cluster was added, less than 10% of the samples would be present within. Although we have optimized the PCA to show the largest differences in the population occasionally the automated clustering method doesn't recognize patterns that are visible upon closer inspection. Additionally, if all samples in your population are very closely related, the clustering (defined by color) may have little meaning. The interpretation of the relationships of samples will require viewing the members of each cluster and their relationship in distance to nearby individuals within and outside their cluster. Finally, one of the strongest measures of the accuracy of the PCA is likely your own knowledge of the relationships of the organisms being sequenced. Using known outgroups within your GBS population will provide a reasonable measure of how much distance on the PCA is equivalent to a sizable genetic distance.
How does PCA work?The PCA method is performed via eigenvalue decomposition of the data covariance or correlation matrix after a data normalization step. Effectively, PCA is a rotation of the axes of the original coordinate system to a new orthongonal axes (principal axes), such that the new axes coincide with the directions of maximum variation for the original observations. More generally, PCA is a mathmatical transformation on a large number of correlated variables into a small number of uncorrelated variables (principal components), i.e. a process of dimensionality reduction. The first principal component accounts for the largest amount of variation possible, the second accounts for less and so on and so forth until all the variance within the dataset is explained. For further information on PCA see the PCA wikipedia page.
5.2 Principal Component Variance
This diagnostic graph displays the variance explained by each of the first 15 Principal Components (PCs 1-15) from Section 5.1. These metrics are valuable because they describe exactly how much variance in the dataset can be explained by each dimension in above 3D PCA. For example: if 75% of the genetic variance detected in your population can be explained by the first 3 principal components, you would have much more confidence in the relationships displayed in the PCA plot than if the first 3 PCs could only explain 25% of the genetic variance. Although no clear cutoffs exist, high levels of dataset variance (>50%) explained in the first 3 PCs inspire much greater confidence in the underlying relationships displayed than low levels of described variance.
5.3 Maximum Likelihood Phylogeny
This phylogenetic tree was assembled by converting VCF data into phylip format with the vcf2phylip program and performing Maximum-likelihood based phylogenetic inference with RAxML. Only SNP sites that had data present for >= 80% of the population were included in this analysis. The ete3 toolkit was used to plot the resulting newick tree file. Each node represents a genetic division in the population and the edge/branch length (noted on each branch) corresponds to the accumulated genetic distance. All data used to produce this phylogeny and the 400 dpi png file can be found in ./VCF/Phylogeny.
6 Genome Imputation
Imputation of Genomic Sites:In addition to the variants discovered by the pipeline, we are providing you with a fully phased and imputed variant call set (./VCF/Imputed_Sites/SNPs.mergedAll.MAFgt0.05.imputed.vcf.gz) that was generated via full genome imputation with the Beagle algorithm (Browning et al. 2007 & Browning et al. 2016). Genotype imputation allows users to accurately fill in the gaps in genotype data inherent to GBS variant calling. Although imputation algorithms can very accurately phase and impute sites, the accuracy of the algorithm is highly dependent on population specific parameters including: genetic relatedness, the length of Identical By Descent (IBD) haplotype blocks, predicted recombination rates and the number of nearby markers considered during imputation. If the population is closely related the IBD haplotype blocks will be larger than if the population is diverse. Imputation algorithms move along each contig looking for sections of shared haplotype in the population and fill in the missing sites with either the variant identified to have a similar haplotype in a reference panel or the highest probability variant based on the population haplotype data and the algorithms statistical model. Before using the imputed markerset for any downstream analyses users are highly encouraged to:
- Read the literature regarding best practices for genotype imputation in their organism of study.
- Evaluate the parameters within ./VCF/Imputed_Sites/SNPs.mergedAll.imputed.log to ensure they conform to the proper standards.
- Perform validation of the imputed SNP sites against a known reference panel.
7 Interpreting Results
Analysis and interpretation of genomic data generated by sequencing technologies are among the most complex problems in genomic sciences. One consequence of this reality is that analysis and interpretation of sequencing data is far from routine. Analytic “best practices” remain very dynamic and reflect our current limited understanding of genomics including technical aspects of data collection.
The process the UW-BRC pipeline uses to analyze your GBS data can be stated as the following:
After VCFs are output from the pipeline, we perform filtering, imputation and generate population analyses (PCA and Phylogenetic Trees) to provide you with insight into the genetic relationships in your GBS population
7.1 Did it work?
Due to the many possible reasons a scientist might have for performing GBS we cannot a priori determine if your experiment was successful for your purposes. However, several metrics may be of interest to you. The first of which is the number of input reads (BarcodedReads) detected per sample. Although no clear rules exist, for a sizable eukaryotic genome (>1 Gb), samples which have in excess of 1x106 reads and low library duplication levels will tend to have good representation of the genotyped sites. Another metric of interest is the total number of SNPs called, and the chromosomes/contigs they are detected on. If you have less SNP sites than you might like on a specific chromosome/contig you should consider the following question. Was the area I wanted to cover reasonably well represented by the enzyme cut site I used in GBS? If not then trying a different enzyme or enzyme combination to cut your genome may yield better representation of that region. If the genome you are using had many contigs and you don't see them in the list, do not worry, for visualization purposes we have only included the top 30 contigs (all SNPs from the missing contigs are still present in your VCFs).
To evaluate how confident you can be in the results, you must go beyond the variant list itself, and evaluate additional aspects of the analyses and perform some basic checks.
8 Credits
8.1 Bioinformatics Resource Center

Bioinformatics Resource Center
Genetics & Biotechnology Center, Room 2130
425 Henry Mall, Madison WI 53706
Email: brc@biotech.wisc.edu
Website: https://bioinformatics.biotech.wisc.edu
8.2 Software
Platform: x86_64-pc-linux-gnu (64-bit)
8.3 References
Browning, S., Browning, B. (2007). Rapid and accurate haplotype phasing and missing data inference for whole genome association studies by use of localized haplotype clustering. Am J Hum Genet, 81, 1084-1097.
Browning, B., Browning, S. (2016). Genotype imputation with millions of reference samples. Am J Hum Genet, 98, 116-126.
Catchen, J., Amores, A., Hohenlohe, P., Cresko, W., Postlethwait, J. (2011) Stacks: Building and Genotyping Loci De Novo From Short-Read Sequences. G3, 1(3),171-182.
Ewing, B., Hillier, L., Wendl, M. C., & Green, P. (1998). Base-Calling of Automated Sequencer Traces Using Phred. I. Accuracy Assessment. Genome Research, 8(3), 175–185. https://doi.org/10.1101/gr.8.3.175
Ewing, B., & Green, P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research, 8(3), 186–194. https://doi.org/10.1101/gr.8.3.186
Glaubitz, C., Casstevens, T., Liu, F., Elshire, R., Sun, Q., & Buckler, E. (2014) TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. Plos One, 9(2):e90346. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0090346
Jiang, H., Lei, R., Ding, S. W., & Zhu, S. (2014). Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics, 15(1), 1–12. https://doi.org/10.1186/1471-2105-15-182
Langmead, B., & Salzberg, S. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9 357–359
Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997.
Li, H., Handsaker, B., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England), 25 (16), 2078–9. https://doi.org/10.1093/bioinformatics/btp352
